Brewer et al., 2019
Ecological and genomic attributes of novel bacterial taxa that thrive in subsurface soil horizons
Brewer, Tess E., Emma L. Aronson, Keshav Arogyaswamy, Sharon A. Billings, Jon K. Botthoff, Ashley N. Campbell, Nicholas C. Dove, Dawson Fairbanks, Rachel E. Gallery, Stephen C. Hart, Jason Kaye, Gary King, Geoffrey Logan, Kathleen A. Lohse, Mia R. Maltz, Emilio Mayorga, Caitlin O’Neill, Sarah M. Owens, Aaron Packman, Jennifer Pett-Ridge, Alain F. Plante, Daniel D. Richter, Whendee L. Silver, Wendy H. Yang, Noah Fierer (2019)
mBio Oct 2019, 10 (5) e01318-19 Cross-CZO National
-
Boulder, GRAD STUDENT
-
Calhoun, Sierra, COLLABORATOR
-
Calhoun, INVESTIGATOR
-
Sierra, GRAD STUDENT
-
Catalina-Jemez, GRAD STUDENT
-
Catalina-Jemez, INVESTIGATOR
-
Sierra, INVESTIGATOR
-
Shale Hills, INVESTIGATOR
-
Reynolds, INVESTIGATOR
-
Sierra, COLLABORATOR
-
National, INVESTIGATOR, STAFF
-
IML, INVESTIGATOR
-
Christina, Luquillo, INVESTIGATOR, COLLABORATOR
-
Calhoun, INVESTIGATOR
-
Luquillo, INVESTIGATOR
-
Boulder, INVESTIGATOR
Abstract
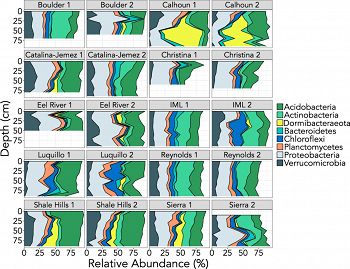
Different soil profiles have distinct microbial communities. Here, we show the relative abundances of the eight most abundant phyla identified from our 16S rRNA gene amplicon data. Not all profiles were sampled to 1 m due to variable bedrock depth. Note that the two profiles sampled from each CZO site were selected to represent distinct soil types (details on soil characteristics are available in Data Set S1 in the supplemental material).
While most bacterial and archaeal taxa living in surface soils remain undescribed, this problem is exacerbated in deeper soils, owing to the unique oligotrophic conditions found in the subsurface. Additionally, previous studies of soil microbiomes have focused almost exclusively on surface soils, even though the microbes living in deeper soils also play critical roles in a wide range of biogeochemical processes. We examined soils collected from 20 distinct profiles across the United States to characterize the bacterial and archaeal communities that live in subsurface soils and to determine whether there are consistent changes in soil microbial communities with depth across a wide range of soil and environmental conditions. We found that bacterial and archaeal diversity generally decreased with depth, as did the degree of similarity of microbial communities to those found in surface horizons. We observed five phyla that consistently increased in relative abundance with depth across our soil profiles: Chloroflexi, Nitrospirae, Euryarchaeota, and candidate phyla GAL15 and Dormibacteraeota (formerly AD3). Leveraging the unusually high abundance of Dormibacteraeota at depth, we assembled genomes representative of this candidate phylum and identified traits that are likely to be beneficial in low-nutrient environments, including the synthesis and storage of carbohydrates, the potential to use carbon monoxide (CO) as a supplemental energy source, and the ability to form spores. Together these attributes likely allow members of the candidate phylum Dormibacteraeota to flourish in deeper soils and provide insight into the survival and growth strategies employed by the microbes that thrive in oligotrophic soil environments.
Importance
Soil profiles are rarely homogeneous. Resource availability and microbial abundances typically decrease with soil depth, but microbes found in deeper horizons are still important components of terrestrial ecosystems. By studying 20 soil profiles across the United States, we documented consistent changes in soil bacterial and archaeal communities with depth. Deeper soils harbored communities distinct from those of the more commonly studied surface horizons. Most notably, we found that the candidate phylum Dormibacteraeota (formerly AD3) was often dominant in subsurface soils, and we used genomes from uncultivated members of this group to identify why these taxa are able to thrive in such resource-limited environments. Simply digging deeper into soil can reveal a surprising number of novel microbes with unique adaptations to oligotrophic subsurface conditions.
Citation
Brewer, Tess E., Emma L. Aronson, Keshav Arogyaswamy, Sharon A. Billings, Jon K. Botthoff, Ashley N. Campbell, Nicholas C. Dove, Dawson Fairbanks, Rachel E. Gallery, Stephen C. Hart, Jason Kaye, Gary King, Geoffrey Logan, Kathleen A. Lohse, Mia R. Maltz, Emilio Mayorga, Caitlin O’Neill, Sarah M. Owens, Aaron Packman, Jennifer Pett-Ridge, Alain F. Plante, Daniel D. Richter, Whendee L. Silver, Wendy H. Yang, Noah Fierer (2019): Ecological and genomic attributes of novel bacterial taxa that thrive in subsurface soil horizons. mBio Oct 2019, 10 (5) e01318-19. DOI: 10.1128/mBio.01318-19
 This Paper/Book acknowledges NSF CZO grant support.
This Paper/Book acknowledges NSF CZO grant support.
Explore Further